My resume is available here: PhuTVan-resume.pdf. If you prefer something more academic and comprehensive, my CV is here: PhuTVan-CV.pdf.
For the more technically inclined, my GitHub repos are located at https://github.com/ptvan.
Brief executive summary
I’m currently the Bioinformatics Solutions Consultant at Pluto Biosciences, supporting our multi-omics computational biology platform. Previously I was the Senior Manager of Bioinformatics Solutions at TwinStrand Biosciences, specializing on duplex sequencing for mutagenesis and cancer applications. Before that I worked as a postdoc, then an analyst at Fred Hutch Cancer Center on flow/mass cytometry and transcriptomics. I received my PhD at Carnegie Mellon where I designed and built a patented fluorescence imager to detect low-abundance proteins. Before grad school I worked on gene transcription networks and proteomics at the Institute for Systems Biology.
My greatest strength is adaptability: in addition to my career in computational biology, I have had success as field ecologist and photojournalist. I am motivated and organized, having worked 3 jobs simultaneously through college and self-published an illustrated children’s book through KickStarter in graduate school.
I enjoy explaining and applying complex ideas. I lectured introductory biology to 200 students and mentored 3 undergraduate teams during my PhD, and my professional life involves collaboration with diverse international researchers. Outside my research day job, I’m a strong advocate for science literacy, mentoring students from elementary school through PhD, and curating an open wiki of data science best practices.
Detailed descriptions of major projects
Multi-omics (2025 - )
I served as the client-facing bioinformatics expert at Pluto Biosciences. I solved problems in ChIPseq/ATACseq, bulk and single-cell RNASeq and advised customers on bioinformatics and statistics. I also worked closely with the engineering team by creating working prototype pipelines to be productionized, collecting Voice of Customers and creating reproducible bug reports.
Genomics (2021 - 2024)
I was the subject matter expert for the computational side of duplex sequencing, connecting the various departments in the company, and overseeing a team of Bioinformatics Scientists. I co-authored papers with our commercial and academic clients, wrote Application Notes and tutorials, and served as second-line expertise for our tech support department.
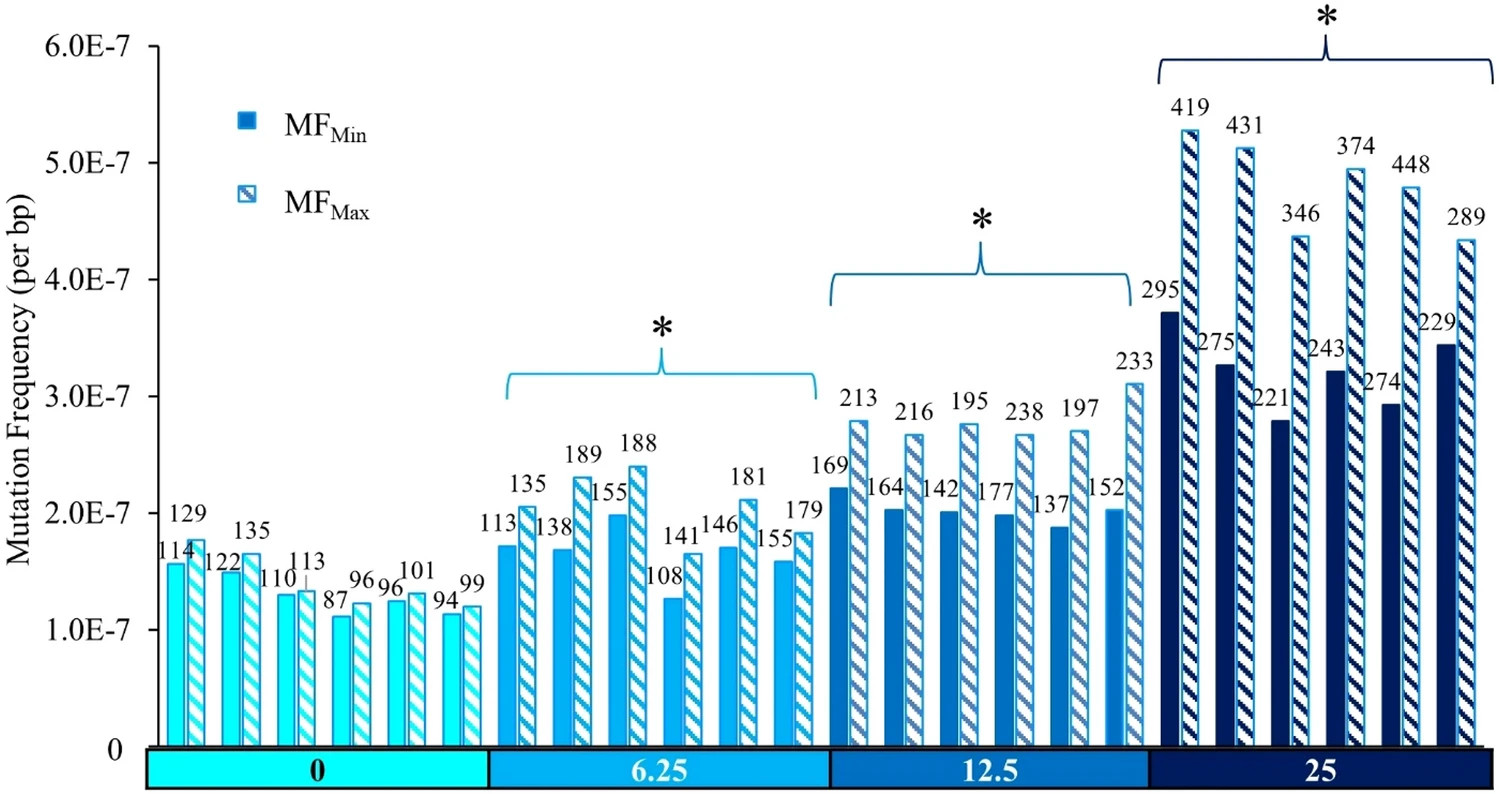
Most recently, I analyzed duplex sequencing data for a Liver-On-Chip (LOC) system, deconvolving mutations from the LOC’s complex mixture of cell types to remove contamination and assess the system’s response to different mutagens. link to paper
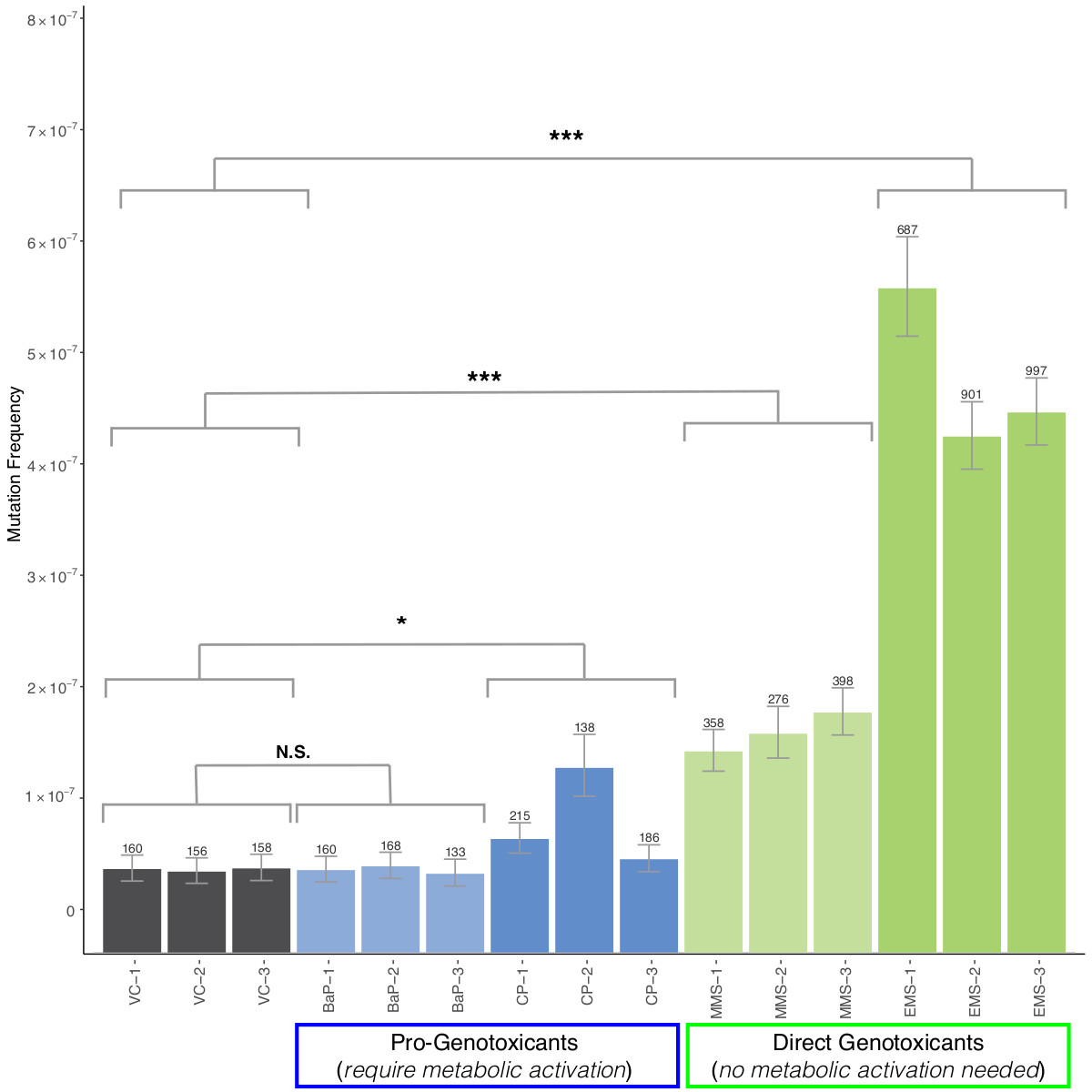
Another project I worked on compared TwinStrand technology to the transgenic rodent assay (TGR) and the alkaline comet assay for detecting mutations induced by NDEA, a common carcinogen link to paper.
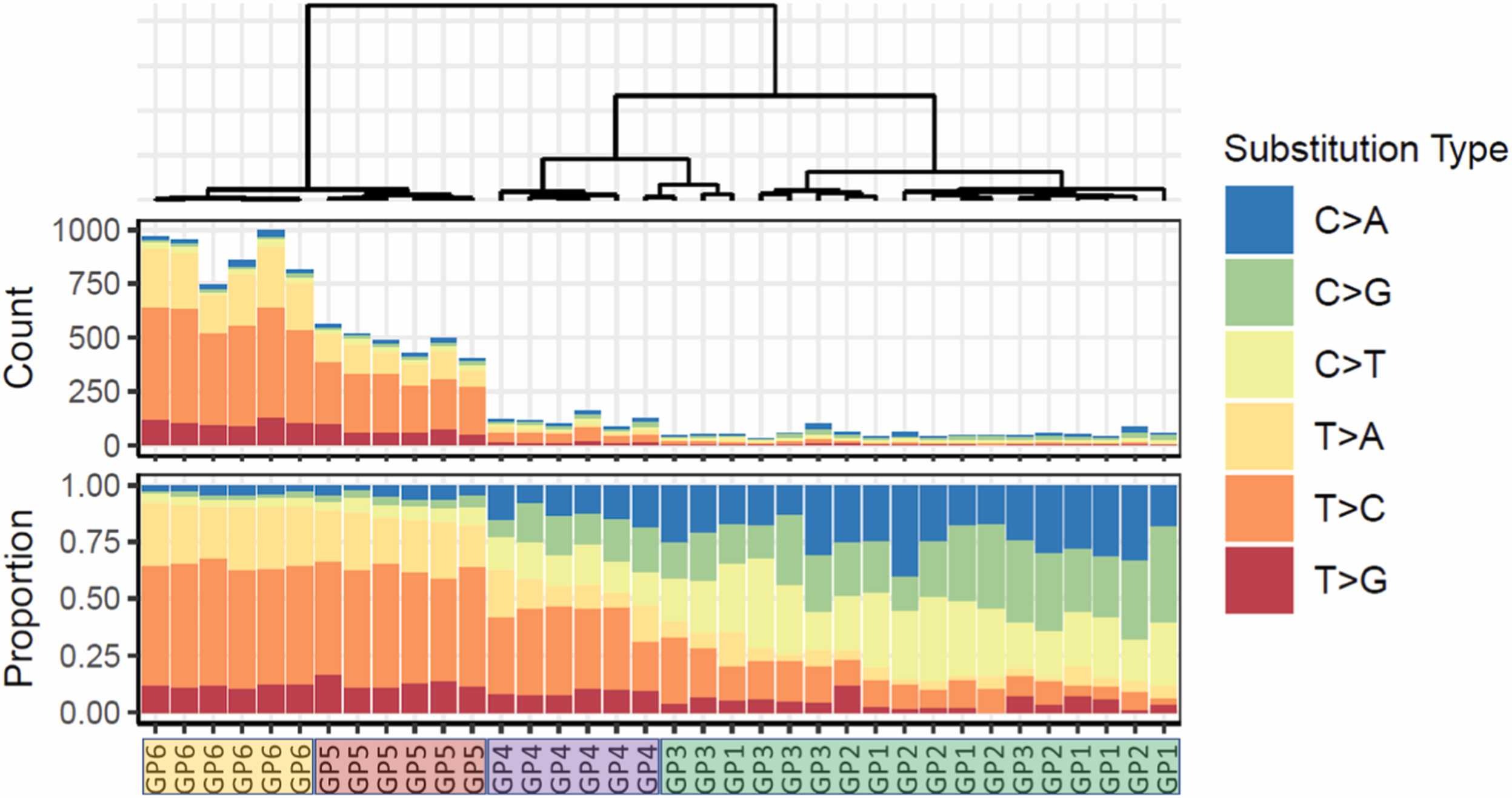
Yet another project I worked on assessed the reproducibility of duplex sequencing across multiple labs link to paper.
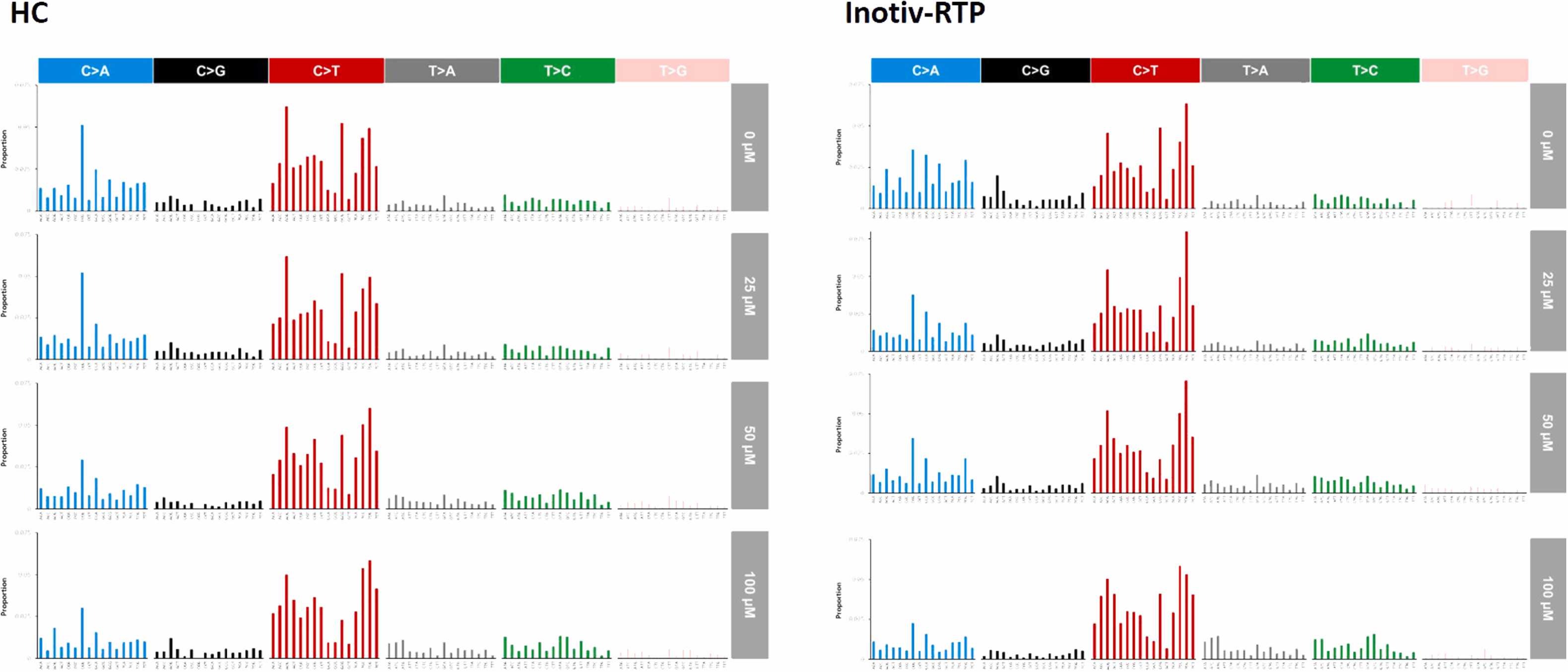
Finally I worked on compared DuplexSeq’s mutagenic detection with the gold-standard LacZ test link to paper.
I started out in the company as a senior Bioinformatics Scientist, analyzing genomic data for commercial and academic clients, going from aligned reads to variants to modeling outcomes. I was also responsible for presenting findings and addressing post-analysis data requests. A year later, I was promoted to lead the newly-formed Bioinformatics Solutions group within the larger Data Sciences Department, managing a team of Bioinformatics Scientists working on cross-functional projects.
Transcriptomics (2017 - 2020)
I was the lead analyst on a project with collaborators at the University of Washington to identify genes that contribute to Tuberculosis resistance in African subjects. I performed data QC, genome alignment, transcript quantification, and downstream analyses (DEG, GSEA, functional annotation, network analysis, etc) link to paper.
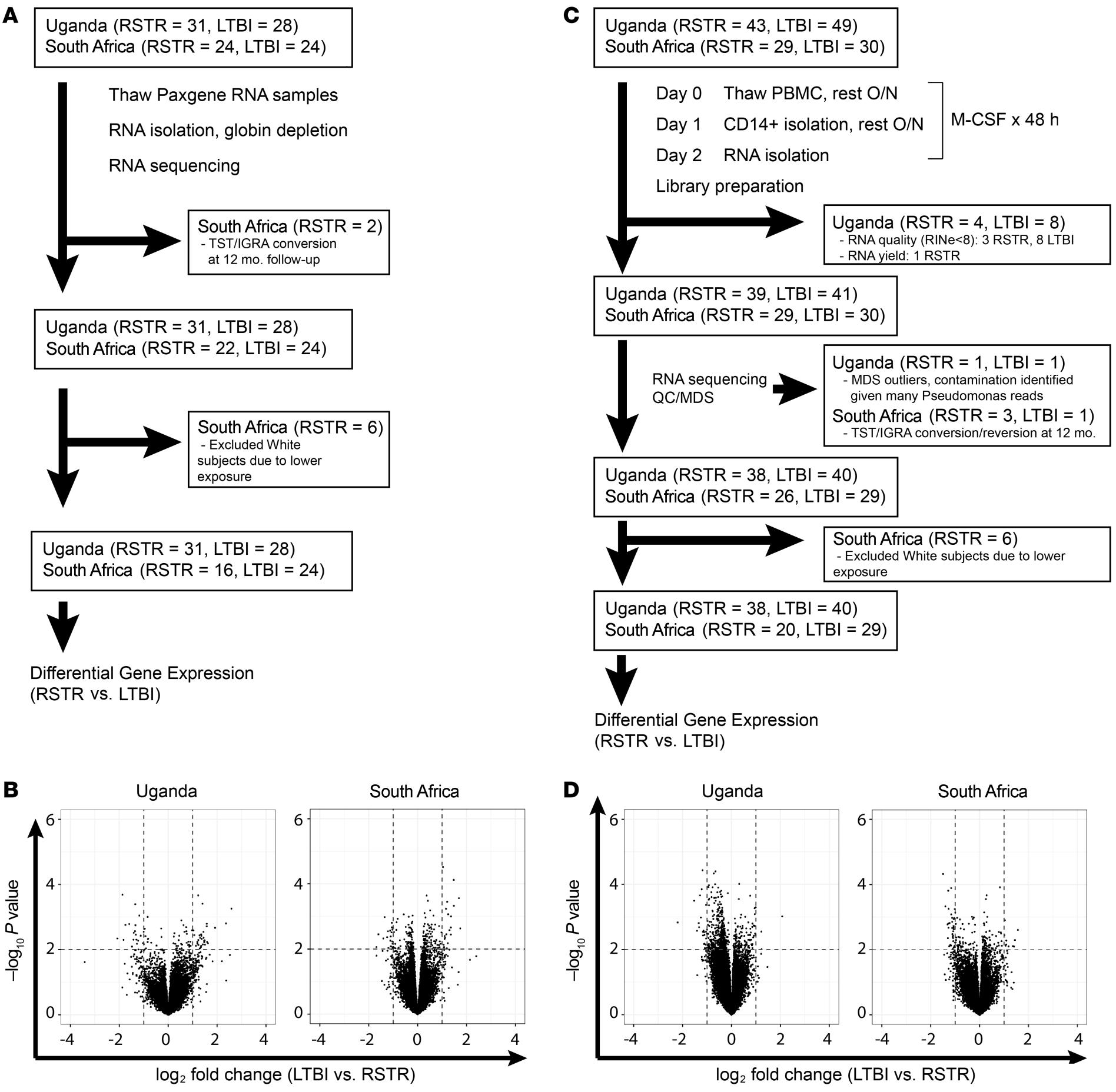
I also contributed code to joint workflows and analyses and was a co-author on a follow-up project on the same cohort. link to paper
My third publication during this period involved some data analysis for the RTS,S/AS01 vaccine for malaria. link to paper.
Another project involved collaborators at the South African Tuberculosis Vaccine Institute to identify possible diagnostic biomarkers for Tuberculosis. I performed my own analyses while also coordinating between Seattle and Cape Town teams in integrating transcriptomic, proteomic and antibody data.
Lastly I also worked on developing a positivity call for Intracellular Cytokine Staining (ICS) data from HIV vaccine trials. The goal of this project is to increase accuracy of the ICS assay while reducing the number of markers required for predicting outcomes.
Flow cytometry & mass cytometry (2014 - 2018)
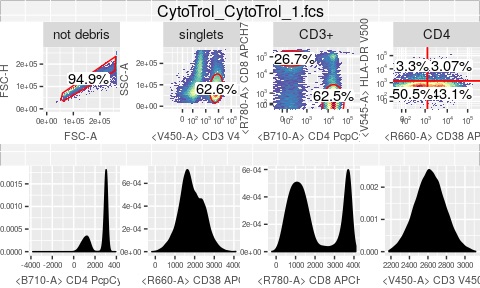
As a postdoc, I helped extend OpenCyto, an open-source R software framework for analyzing high-dimensional flow-cytometry and mass-cytometry data. I also worked on ggCyto, an R package that enables ggplot-style plotting of flow- and mass-cytometry datasets. link to paper
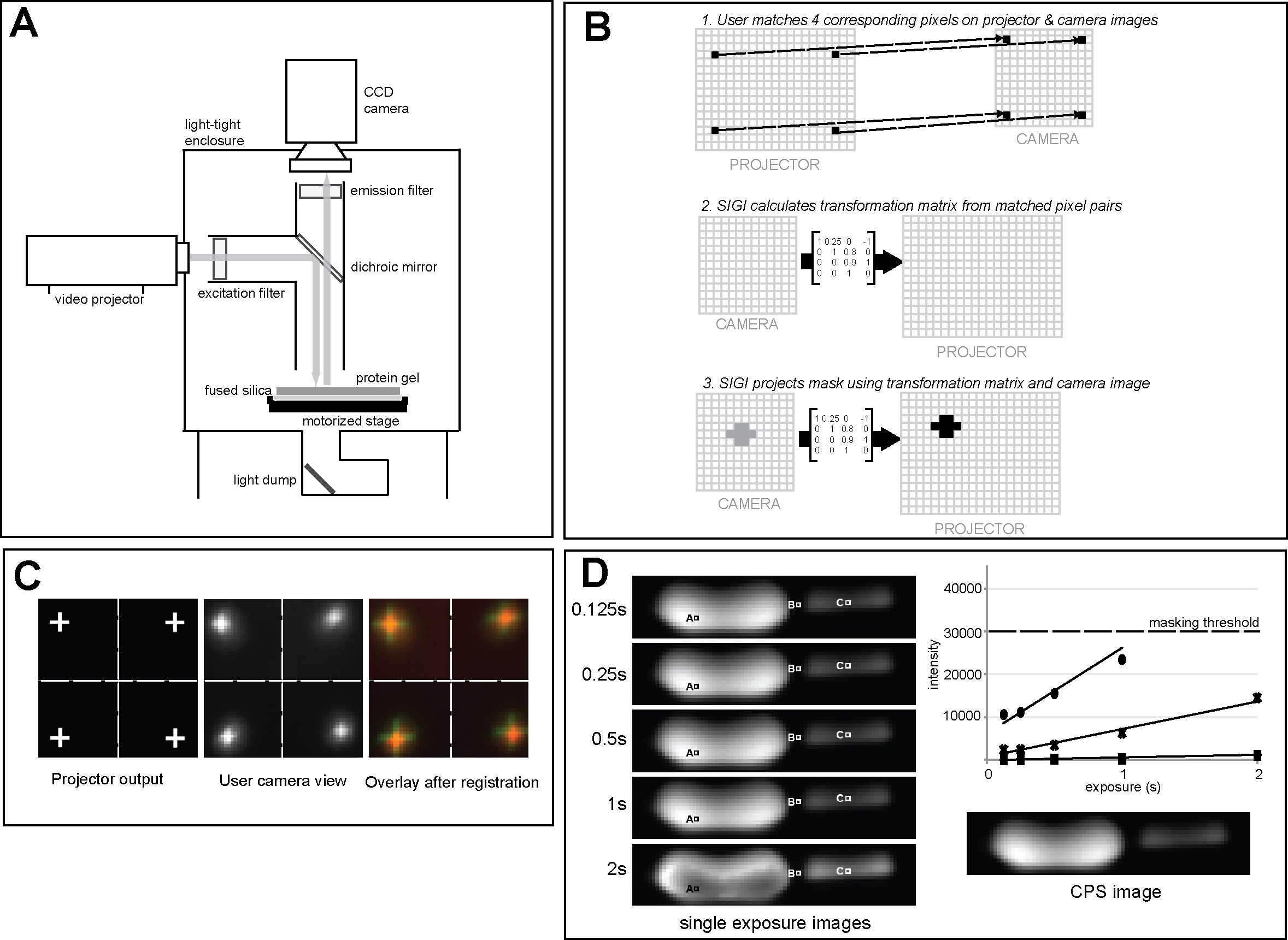
Structured Illumination Gel Imager (SIGI) and 2DE proteomics (2009 - 2014)
My PhD contained 3 parts: first, I built a high-dynamic-range imager to detect rare proteins in 2-dimensional electrophoretic (2DE) gels, which we dubbed SIGI. SIGI captured multiple exposures of the 2DE gels containing fluorescently-labelled proteins with structured illumination from an LCD projector and automatically assembled the final 32-bit grayscale images. SIGI also contained a robotic cutting arm that can excise the proteins from the gels for sequencing by tandem mass spectra (MS/MS). Carnegie Mellon was granted a US patent on SIGI after I graduated. link to paper
Second, I developed and refined an agarose stacking gel that improved protein retention during the preparation of 2DE gels for MS/MS sequencing. link to paper
Lastly, I mentored three 2-person teams of CMU undergrads in preparing 2DE protein gels, operating the imager and performing data analysis on their experiments.
Microbial oxidative stress response network (2008 - 2009)
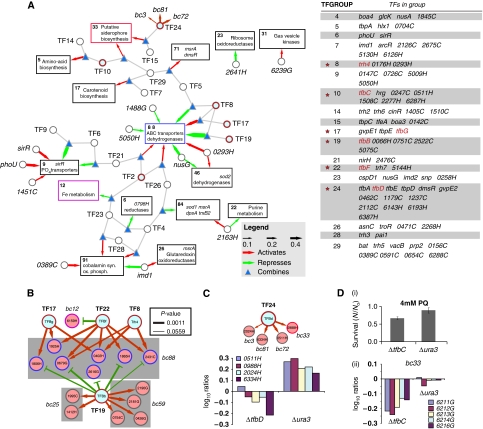
Working at the Institute for Systems Biology (ISB) as a research associate, I worked on an extension for Inferelator, an algorithm for predicting regulators of gene expression, implemented in R at the time. This branch of the algorithm ended up being superseded, but you can see what I worked on in this Github repository. This work enabled us to build and test a model of oxidative stress response in the archaeon Halobacterium salinarum, which can survive extremely salty environments like Utah’s Great Salt Lake. link to paper
Microbial PeptideAtlas database and web portal (2006 - 2008)
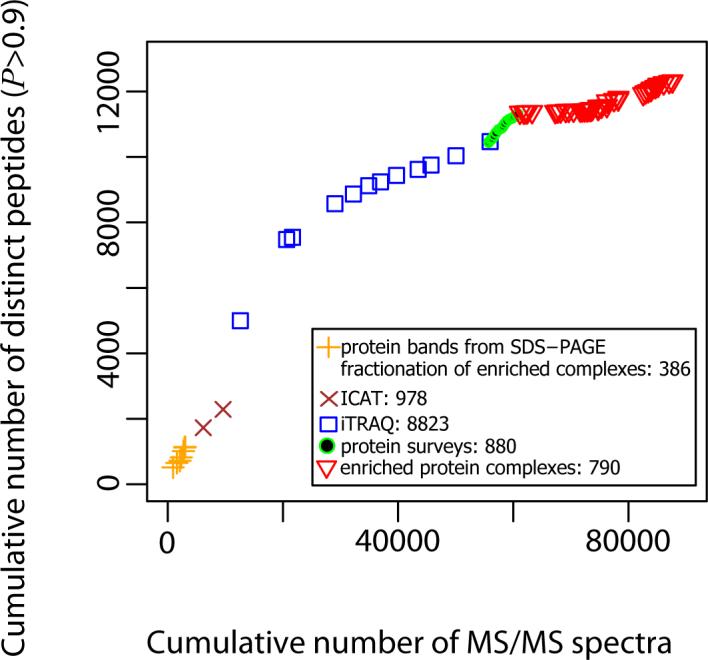
During my internship at ISB, I converted peptide mass spectra of Halobacterium experiments from vendor binary formats into mzXML, mapped spectra to peptides then loaded the proteins into SQLServer-backed web portal. We also found biases in peptide detection depending on the sequencing method used. link to paper
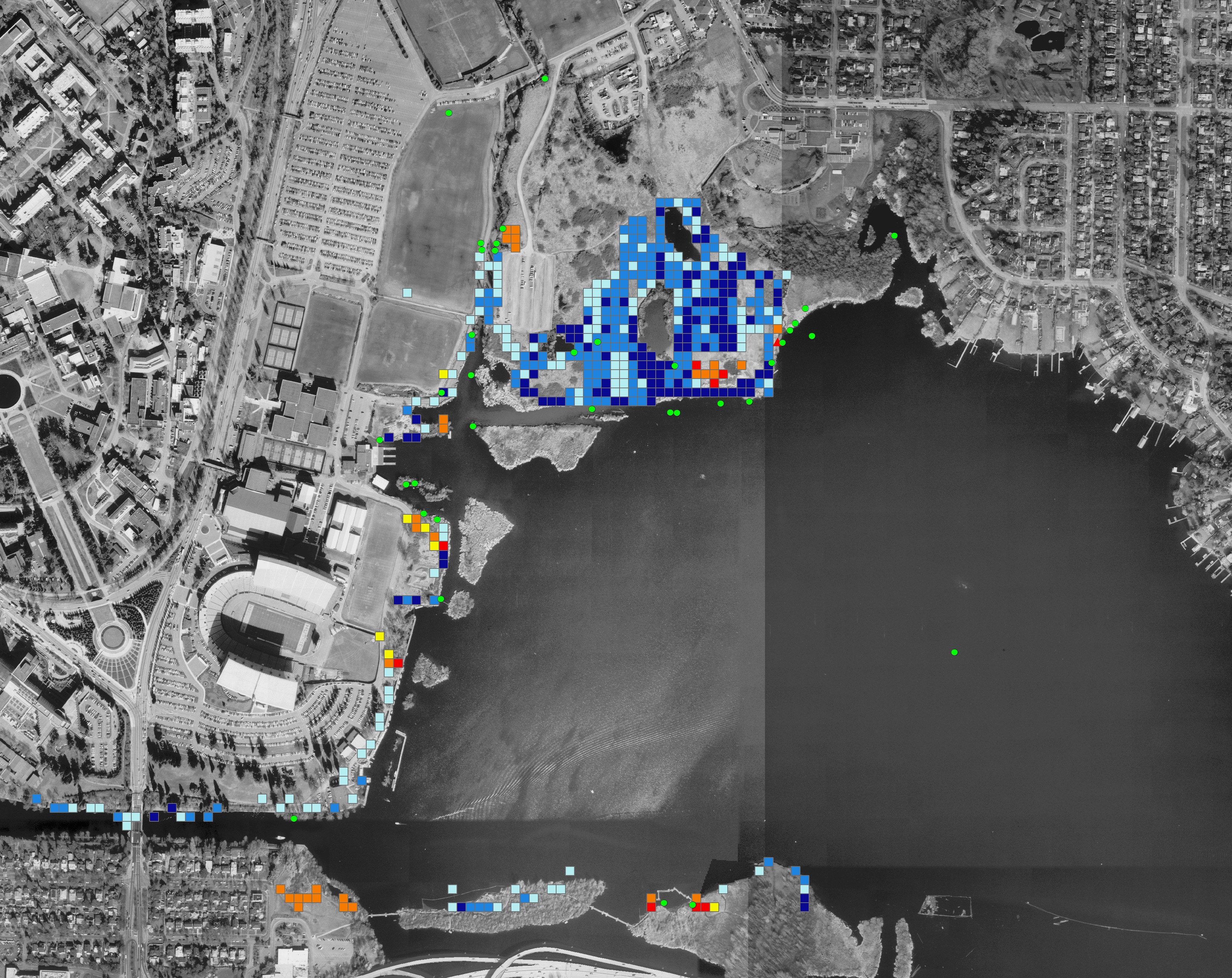
Geospatial model of rodent spread in Seattle (2006 - 2007)
As part of our Bachelor theses, my University of Washington classmate and good friend Filip and I collected sightings of the rodent nutria (Myocastor coypus) in western Washington, mainly in the area surrounding Union Bay. I created a linear model to predict the spread of the species in Seattle using R and ArcGIS. This project ended up informing a decision by the UW’s Environmental Health and Safety not to undertake eradication efforts, since much of the nutria population was not surviving through the winters anyway.